| Amygdalin inhibits genes related to cell cycle in SNU-C4 human colon cancer cells |
| 发布时间:2012-07-24 信息来源:admin 发布人:admin 点击次数:4149 |
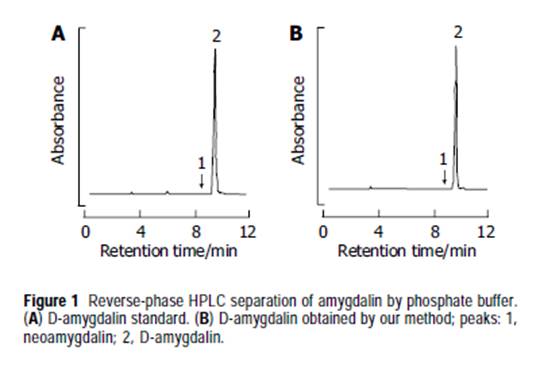
Abstract AIM: The genes were divided into seven categories according to biological function; apoptosis-related, immune response-related, signal transduction-related, cell cyclerelated, cell growth-related, stress response-related and transcription-related genes. METHODS: We compared the gene expression profiles of SNU-C4 cells between amygdalin-treated (5 mg/mL,24 h) and non-treated groups using cDNA microarray analysis. We selected genes downregulated in cDNA microarray and investigated mRNA levels of the genes by RT-PCR. RESULTS: Microarray showed that amygdalin downregulated especially genes belonging to cell cycle category: exonuclease 1 (EXO1), ATP-binding cassette, sub-family F, member 2 (ABCF2), MRE11 meiotic recombination 11 homolog A (MRE11A), topoisomerase (DNA) I (TOP1), and FK506 binding protein 12-rapamycin-associated protein 1 (FRAP1). RT-PCR analysis revealed that mRNA levels of these genes were also decreased by amygdalin treatment in SNU-C4 human colon cancer cells. CONCLUSION: These results suggest that amygdalin have an anticancer effect via downregulation of cell cycle-related genes in SNU-C4 human colon cancer cells, and might be used for therapeutic anticancer drug. Key words: Amygdalin; SNU-C4; cDNA microarray; Cell cycle INTRODUCTION The incidence of colorectal cancer has been increasing rapidly since 1975[1] with about 300 000 new cases and 200 000 deaths in Europe and USA every year[2,3]. Death from colorectal cancer is the second commonest cause of any cancer in men in the European Union[4]. The stage of the disease is an important effect to long-term survival of colorectal cancer. If detected early, it may be curable by surgery. But once metastases develop the prognosis becomes poor. At least 40% of patients with colorectal cancer develop metastases during their illness[5]. Although various therapies as surgery, radiotherapy, and chemotherapy are used on advanced cancer, the most effective approach is yet to be discovered. Amygdalin is ingredient of Prunus persica Batsch (Persicae semen, Rosaceae), Prunus armeniaca L. var. ansu Max (Armenicae semen, Rosaceae) and Prunus amygdalus Batsch var. amara (Amygdali semen amara, Rosaceae), and these are abundant in the seeds of bitter almond and apricots[6]. The evidence for effect of amygdalin has been reviewed as prevention and control of cancers. Due to cyanide toxicity, there has been controversy for amygdalin as cancer drug[7-9]. However, Moertel et al., demonstrated that, in human, intravenous infusion of amygdalin produced neither cyanidemia nor signs of toxicity but that oral administration resulted in significant blood cyanide levels and that, in one case, oral amygdalin plus almond extract produced transient symptoms of cyanide intoxication and further increase of blood cyanide[9]. Additionally, Fukuda et al.[10], reported anti-tumor effect of amygdalin and other components of Prunus persica seeds. Kwon et al.[13], reported that controversy on anticancer effect of amygdalin was due to its conversion to inactivate isoform, and Persicae semen extract induced apoptosis. Although taking a growing interest and reports for amygdalin as cancer drug, anti-cancer mechanism of amygdalin has not been reported. In this study, we determined whether amygdalin-treated SNU-C4 cells were susceptible to cell death, and compared the expression profiles of SNU-C4 cells between amygdalintreated group and non-treated group using cDNA microarray analysis. We also performed RT-PCR for genes selected by cDNA microarray analysis. MATERIALS AND METHODS Preparation of amygdalin Both 500 g of Armeniacae semen hatched from the shell and 10 L of 4% citric acid solution were refluxed for 2 h. Filtered when it was still hot, the filtrate was passed through the column packed with HP-20. The substance absorbed within the column was concentrated after it had been eluted by ethanol. 4.2 g of amygdalin (with the yield rate of 0.84%) was obtained by recrystallizing the extract with ethanol. The amygdalin was used after it had been determined to be over 95.0% of purity, by means of high-pressure liquid chromatography (HPLC) to measure its purity (Figure 1).
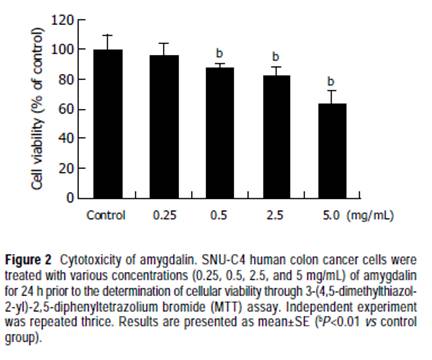
Cell culture The SNU-C4 cell line was obtained from the Korean Cell Line Bank (KCLB, Seoul, South Korea). Cells were cultured in RPMI-1640 medium (Gibco, NY, USA) supplemented with 10% heat-inactivated FBS (Gibco). Cultures were maintained in a humidified incubator at 37 in an atmosphere of 50 mL/L CO2, 95% air, and the medium was changed every 2 d. MTT assay Cell viability was determined by 3-(4,5-dimethylthiazol-2-yl) -2,5-diphenyltetrazolium bromide (MTT) assay. Cells were seeded in triplicate at a concentration of 1×105 cells/well on a 96-well plate. SNU-C4 cells were treated with amygdalin at concentrations of 0.25, 0.5, 2.5, and 5 mg/mL for 24 h.After MTT (Sigma, MO, USA) was added to each group,the cells were incubated for 4 h. Then, they were further incubated for 1 h, including the solution in which MTT was dissolved. The viability was measured with a microtiter plate reader (Bio-Tek, VT, USA) at a test wavelength of 595 nm with a reference wavelength of 690 nm. The optical density (A) was calculated as the difference between the reference wavelength and the test wavelength. Percent viability was calculated as (A of drug-treated sample/A of untreated sample)×100. cDNA synthesis Total RNA was extracted using RNAzolTM B (TEL-TEST,TX, USA) as per the manufacturer’s protocol. cDNA synthesis was performed with 3DNATM Array 50TM detection method (Genisphere, PA, USA) as per the manufacturer’s protocols. In the control and amygdalin treatment (5 mg/mL,24 h), cDNA was synthesized from total RNA as follows:3 μL of RT primer, total RNA and additional nuclease-free water were mixed to form a 29 μL RNA-RT primer mix,which was microfuged briefly, heated to 80 for 10 min,and immediately transferred to ice. One microliter of the RNase inhibitor Superase-InTM was added to the RNA-RT primer mix. Eight microliters of 5× SuperScript II First Strand Buffer (Gibco), 2 μL of dNTP mix (10 mmol/L each of dATP, dCTP, dGTP, dTTP), 4 μL of 0.1 mol/L dithiothreitol, 2 μL of Superscript II enzyme (400 units), and 3 μL of RNase-free water were mixed in each microtube, and the RNA-RT primer mix was then added. The tubes were then incubated at 42 for 2 h, and the reaction was heated by adding 7 μL of 0.5 mol/L NaOH/50 mmol/L EDTA. The microtubes were then incubated for denaturation at 65 for 10 min, and neutralization was carried out by adding 10 μL of 1 mol/L Tris-HCl at pH 7.5. The contents of two tubes were combined to yield a 130 μL cDNA solution in one single tube. The original tubes were rinsed with 16 μL of 10 mmol/L Tris at pH 8.0/1 mmol/L EDTA. Upon completion of the synthesis procedure, the cDNA solution was concentrated by ethanol precipitation. Three microliters of thoroughly vortexed 5 mg/mL linear acrylamide solution was added to the cDNA solution. Six microliters of NaCl and 540 μL of 95-100% ethanol was then added and moderately vortexed. The mixture was then incubated at -20 for 30 min, centrifuged at >10 000 g for 15 min, and the supernatant was aspirated. The cDNA pellet was washed with 300 μL of 70% ethanol. After centrifuging again at >10 000 g for 5 min, the supernatant was aspirated, and the cDNA pellet was completely dried at 65 over a period of 10-30 min. Microarray hybridization A cDNA chip of TwinChipTM Human 8K (Digital Genomics, Seoul, South Korea) was used. The concentrated cDNA and 3DNATM were hybridized on two identical arrays in a slide for a duplicate experiment. Two times formamide-based hybridization buffer was thawed and responded by heating at 55 for 10 min with intermittent inversions, and then microfuged for 1 min. Ten microliters of nuclease-free water was added to the cDNA pellet, and the cDNA was completely resuspended by heating at 55 for 10 min with intermittent inversions, and then microfuged for 1 min. Ten microliters of nuclease-free water was added to the cDNA pellet, and the cDNA was completely resuspended by heating at 65 for 10-15 min and vortexing for 5 min. Thirty microliters of hybridization mixture was prepared from 10 μL of cDNA, 15 μL of 2× hybridization buffer, 2 μL of Array50 dT Blocker, and 3 μL of nuclease-free water. The hybridization mixture was incubated at 80 for 10 min and then at 50 for 30-60 min. The hybridization mix was then added to the pre-warmed chip. After a disposable coverslip was applied, the chip was incubated overnight in a dark humidified chamber at 50 . After serial washing, the array was incubated for 2 min at room temperature with 95% ethanol. The slide was immediately transferred to a dry 50-mL centrifuge tube and dried by centrifugation for 2 min at 800-1 000 r/min. Array50 capture reagent was then thawed in the dark at room temperature over a period of 20 min and vortexed for 3 s. Thirty microliters of hybridization mixture was prepared from 15 μL of 2× hybridization buffer, 2.5 μL of 3DNA TM capture reagent #1 (Cy3), 2.5 μL of 3DNA TM capture reagent #2 (Cy5), and 10 μL of nuclease-free water. Following gentle vortexing and brief microfuging, the hybridization mixture was incubated at 80 for 10 min and then at 50 for 20 min. The hybridization mixture was applied to the pre-warmed chip after it was removed from the incubator. After a disposable coverslip was applied, the microarray was incubated in a dark humidified chamber at 50 for 2-3 h. After serial washing, the slide was immediately transferred to a dry 50-mL centrifuge tube and dried by centrifugation for 2 min at 800-1 000 r/min, and then transferred to a dark slide box. Scanning and data analysis The hybridized microarray was scanned with a confocal laser scanning microscope (ScanArray 5000; Packard Inc.,CT, USA) at 532 nm for Cy3 and 635 nm for Cy5. Image analysis using GenePix (Axon Inc., CA, USA) produced quantitative values for each microarray spot. Pixel intensity of the background was subtracted from those of microarray spots. Spot intensities were normalized using the intensities generated by intensity/location dependent method[11].Normalized spot intensities were calculated into gene expression ratios between the control and treatment groups.Mean data acquired from two identical arrays in a single slide of TwinChipTM were analyzed. RT-PCR Two micrograms of sample of RNA isolated from SNU-C4 cells using RNAzolTM B (TEL-TEST) and 2 μL of random hexamers (Promega, WI, USA) were added together, and the mixture was heated at 65 for 10 min. AMV reverse transcriptase (Promega) of 1 μL, 5 μL of 10 mmol/L dNTP (Promega), 1 μL of RNasin (Promega) and 5 μL of 10× AMV RT buffer (Promega) were then added to the mixture,and the final volume was brought up to 50 μL with diethyl pyrocarbonate-treated water. Subsequent PCR amplification was performed in a reaction volume of 40 μL containing 1 μL of the appropriate cDNA, 1 μL of each set of primers at a concentration of 10 pmol/L, 4 μL of 10× reaction buffer, 1 μL of 2.5 mmol/L dNTP and 2 U of Taq DNA polymerase (TaKaRa Bio Inc., Shiga, Japan). We selected five genes, exonuclease 1 (EXO1), ATP-binding cassette (ABC), sub-family F, member 2 (ABCF2), MRE11 meiotic recombination 11 homolog A (MRE11A), topoisomerase (DNA) I (TOP1), and FK506 binding protein 12-rapamycin associated protein 1 (FRAP1) downregulated in microarray analysis by treatment of amygdalin and performed PCR. Primer sequences, annealing temperatures and products size of genes are summarized in Table 1. The RT-PCR products were electrophoresed on a 1.5% agarose gel and visualized by staining with ethidium bromide. Statistical analysis Results were expressed as mean±SE. The data were analyzed by one-way ANOVA following the Dunnett’s post hoc analysis, using SPSS. Differences were considered significant at P<0.05. RESULTS Cytotoxicity of amygdalin in SNU-C4 cells When treated with amygdalin of 0.25, 0.5, 2.5, and 5 mg/mL concentrations for 24 h, viabilities of SNU-C4 cells were 96.1±7.4%, 87.8±2.4%, 82.9±5.5%, and 63.7±8.3%,respectively, compared with those of non-treated cells (Figure 2), and MTT assay showed dose-dependent cytotoxicity of amygdalin on SNU-C4 cells. Therefore, to compare the precise effect of amygdalin, further experiments were carried out, using 5 mg/mL amygdalin.
Analysis of microarray expression data In order to assess the expression profiles in SNU-C4 cells treated with amygdalin (5 mg/mL, 24 h), cDNA microarray that contained duplicate cDNA probes from 8k human clones was performed to be screened at a time. Figure 3 showed the image of 8k human cDNA microarray. The green spots represented genes of control group that was labeled by Cy3-captured reagent and overexpressed than amygdalin group. In contrast, the red spots represented genes of amygdalin group that was labeled by Cy5-captured reagent and were overexpressed than control. The yellow spots represented genes that showed no difference in expression level between control and amygdalin group.
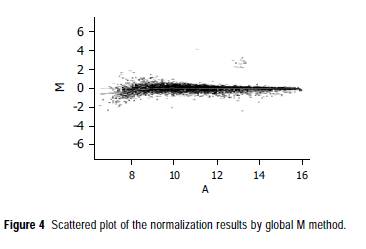
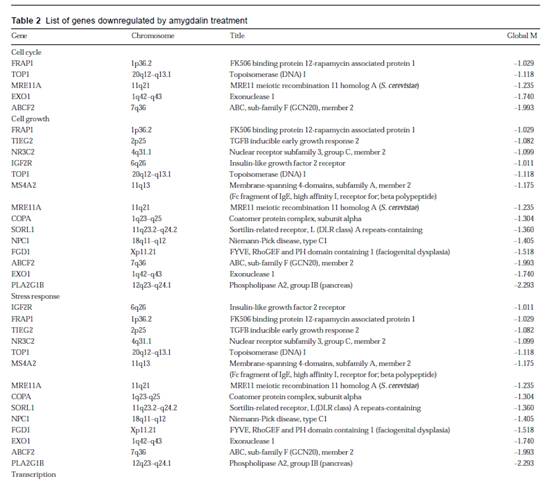
To normalize intensity ratio of each gene expression pattern, global M method was used in this study. First, the primary data were normalized by the total spots of intensity between two groups, and then normalized by the intensity ratio of reference genes, such as housekeeping genes in both groups. Finally, the expression ratio of control and amygdalin group was converted to log2 ratio of each gene, and was represented by a scattered plot in Figure 4. After normalizing the data, a difference in the normalized intensity ratio was selected. The genes of expression ratio were observed to be lower than -1 for which the observed expression was downregulated by amygdalin (Table 2). The genes downregulated by amygdalin were divided into seven categories by biological function; apoptosis-related gene, response to immune-related gene, signal transduction-related gene, cell cycle-related gene, cell growth-related gene, response to stress-related gene and transcription-related gene. We paid attention to genes belonging to cell cycle category. EXO1, ABCF2, MRE11A, TOP1 and FRAP1 cannot belong to category of cell cycle but category of cell growth, stress response, and transcription.
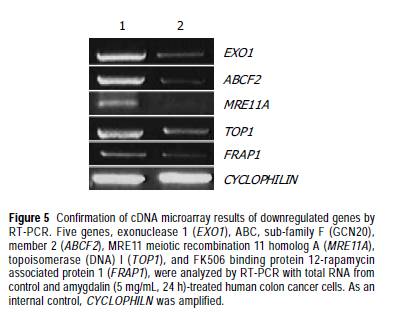
Confirmation of cDNA microarray findings by RT-PCR We selected EXO1, ABCF2, MRE11A, TOP1, and FRAP1 among the downregulated genes by amygdalin treatment, and observed mRNA expressions of EXO1, ABCF2, MRE11A, TOP1, and FRAP1 using RT-PCR that reproduced the results of cDNA microarray. The efficiency of the reaction was adjusted by CYCLOPHILIN amplification. As shown in Figure 5, the expressions of EXO1, ABCF2, MRE11A, TOP1, and FRAP1 were decreased by amygdalin treatment (5 mg/mL, 24 h).
DISCUSSION Amygdalin belongs to a family of compounds called cyanogenic glycosides. Cyanide is believed to be the active cancer-killing ingredient in amygdalin. Amygdalin would be broken down by an enzyme in cancerous tissue, and toxic
cyanide to release from broken amygdalin would kill the cancer. It is further hypothesized that another enzyme, rhodanese, which has the ability to detoxify cyanide, is present in normal tissues but deficient in cancer cells. These two factors supposedly combine to effect a selective poisoning of cancer cells by the cyanide, while normal cells remain undamaged[6,12]. However, amygdalin is not approved by the Food and Drug Administration (FDA) for use in USA, and the anticancer effect of amygdalin has admitted controversy. Kwon et al., demonstrated that the controversy on amygdalin was due to its conversion to inactivate isoform, and Persicae Semen extract induced apoptosis in human promyelocytic leukemia (HL-60) cells with IC50 of 6.4mg/mL in the presence of 250 nmol/L of β-galactosidase[13]. One of the advantages of cDNA microarray is the possibility to observe the expression pattern of the whole genes and compare with different conditions. This study is the first work of comparison of the gene expression profiles of SNU-C4 human colon cancer cells after amygdalin treatment. cDNA microarray analysis showed that amygdalin treatment downregulated genes belong to categories of cell cycle-related gene, and related to cell growth, response to stress and transcription. Anticancer agents targeting DNA and DNA-associated processes are widely used in the treatment of human cancers and produce significant increases in the survival of patients[14]. In general, four complex systems have evolved to respond to DNA damage: DNA repair, cell cycle checkpoint control, apoptosis, and damage tolerance[15]. EXO1 is involved in mismatch repair and recombination, and has been considered as a candidate gene for colorectal susceptibility[16,17]. ABCF2 is a member of the superfamily of ABC transporters. ABC proteins transport various molecules across extra- and intracellular membranes[18]. MRE11A encodes a nuclear protein involved in homologous recombination, telomere length maintenance, and DNA double-strand break repair[19,20]. TOP1 acts as a swivel during replication, transcription, and recombination to relieve overwinding if duplex DNA[15,21]. DNA topoisomerase inhibitors represent a major class of anticancer agents (e.g. camptothecin; CPT) with documented activities against a broad spectrum of human malignancies[22]. FRAP1 has been identified as the downstream target of the FKBP12-rapamycin complex, which inhibits progression through G1 of the cell cycle[23], and mediates cellular responses to stresses such as DNA damage and nutrient deprivation[24,25]. EXO1, ABCF2, MRE11A, TOP1, and FRAP1 particularly belong to categories of cell cycle-related gene and associated to DNA replication, repair or recombination. Thus, downregulation of these genes by amygdalin would induce DNA damage in SNU-C4 cells. In confirmation through RT-PCR, mRNA expressions of these five genes were also decreased by amygdalin treatment. Our results indicate that amygdalin induced DNA damage, involved in cell cycle on SNU-C4 human colon cancer cells and amygdalin might be used for therapeutic anticancer drug. REFERENCES 1 Singh P, Dai B, Dhruva B, Widen SG. Episomal expression of sense and antisense insulin-like growth factor (IGF)-binding protein-4 complementary DNA alters the mitogenic response of a human colon cancer cell line (HT-29) by mechanisms that are independent of and dependent upon IGF-I. Cancer Res 1994; 54: 6563-6570 2 Midgley R, Kerr D. Colorectal cancer. Lancet 1999; 353: 391–399 3 Kim EJ, Schaffer BS, Kang YH, MacDonald RG, Park JH. Decreased production of insulin-like growth factor-binding protein (IGFBP)-6 by transfection of colon cancer cells with an antisense IGFBP-6 cDNA construct leads to stimulation of cell proliferation. J Gastroenterol Hepatol 2002; 17: 563–570 4 Boyle P, Langman JS. ABC of colorectal cancer: epidemiology. BMJ 2000; 321: 805-808 5 Szepeshazi K, Schally AV, Groot K, Armatis P, Halmos G, Herbert F, Szende B, Varga JL, Zarandi M. Antagonists of growth hormone-releasing hormone (GH-RH) inhibit IGF-II production and growth of HT-29 human colon cancers. Br J Cancer 2000; 82: 1724-1731 6 Cho CJ, Yang HS, Kim TH, Chong SY, Lee IR, Kim JH, Leem JP, Chong MH, Lee MW, Lee KY, Pae KH, Kim JW, Han DR. Hyundae-Saengyak-Hak, 8th ed. Hak Chang Press 1998: 321-323 7 Flora KP, Cradock JC, Ames MM. A simple method for the estimation of amygdalin in the urine. Res Commun Chem Pathol Pharmacol 1978; 20: 367-378 8 Khandekar JD, Edelman H. Studies of amygdalin (laetrile) toxicity in rodents. JAMA 1979; 242: 169-171 9 Moertel CG, Ames MM, Kovach JS, Moyer TP, Rubin JR, Tinker JH. A pharmacologic and toxicological study of amygdalin. JAMA 1981; 245: 591-594 10 Fukuda T, Ito H, Mukainaka T, Tokuda H, Nishino H Yoshida T. Anti-tumor promoting effect of glycosides from Prunus persica seeds. Biol Pharm Bull 2003; 26: 271-273 11 Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 2002; 30: e15 12 Newmark J, Brady RO, Grimley PM, Gal AE, Waller SG, Thistlethwaite JR. Amygdalin (Laetrile) and prunasin betaglucosidases: distribution in germ-free rat and in human tumor tissue. Proc Natl Acad Sci USA 1981; 78: 6513-6516 13 Kwon HY, Hong SP, Hahn DH, Kim JH. Apoptosis induction of Persicae Semen extract in human promyelocytic leukemia (HL-60) cells. Arch Pharm Res 2003; 26: 157-161 14 Capranico G, Zagotto G, Palumbo M. Development of DNA topoisomerase-related therapeutics: a short perspective of new challenges. Curr Med Chem Anti Canc Agents 2004; 4:335-345 15 Wang Z. DNA damage-induced mutagenesis: a novel target for cancer prevention. Mol Interv 2001; 1: 269-281 16 Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res 1998; 58: 5027- 5031 17 Wilson DM 3rd, Carney JP, Coleman MA, Adamson AW, Christensen M, Lamerdin JE. Hex1: a new human Rad2 nuclease family member with homology to yeast exonuclease 1.Nucleic Acids Res 1998; 26: 3762-3768 18 Dean M, Hamon Y, Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res 2001; 42:1007-1017 19 Paull TT, Gellert M. The 3’ to 5’ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell 1998; 1: 969-979 20 Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem 1998; 273: 21447-21450 21 Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 2001; 70: 369-413 22 Liu LF. DNA topoisomerase poison as antitumor drugs. Ann Rev Biochem 1989; 58: 351-375 23 Astier AL, Xu R, Svoboda M, Hinds E, Munoz O, de Beaumont R, Crean CD, Gabig T, Freedman AS. Temporal gene expression profile of human precursor B leukemia cells induced by adhesion receptor: identification of pathways regulating B-cell survival. Blood 2003; 101: 1118-1127 24 Brown EJ, Albers MW, Bum Shin T, Ichikawa K, Keith CT, Lane WS, Schreiber SL. Amammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994; 369: 756-758 25 Chiu MI, Katz H, Berlin V. RAPT1, a mammalian homology of yeast Tor, interacts with the FKBP12/rapamycincomplex. Proc Natl Acad Sci USA 1994; 91: 12547-12578 |







 Amygdalin inhibits genes related to cell cycle in SNU-C4 human colon cancer cells
Amygdalin inhibits genes related to cell cycle in SNU-C4 human colon cancer cells